At the time of the writing of this post, the Biodiversity Information Standards conference, TDWG 2015, is on in Kenya and everyone around the world can listen to the live audio stream. Data sharing, data re-use, and data discovery are being brought up in almost every talk. We might have entered the age of big data twenty years ago, but it is now that scientists face the real challenge – storing and searching through the deluge of data to find what they need.
As the rate at which we exponentially generate data exceeds the rate at which data storage technologies improve, the field of data management seems to be greatly challenged. Worse, this means the more new data is generated, the more of the older ones will be lost. In order to know what to keep and what to delete, we need to describe the data as much as possible, and judge the importance of datasets. This post is about a novel way to automatically generate scientific papers describing a dataset, which will be referred to as data papers.
The common characters of the records, i.e. descriptions of the object of study, the measurement apparatus and the statistical summaries used to quantify the records, the personal notes of the researcher, and so on are called metadata. Major web portals such as DataONE or the Global Biodiversity Information Facility store metadata in conjunction with a given dataset as one or more text files, usually structured in special formats enabling the parsing of the metadata by algorithms.
To make the metadata and the corresponding datasets discoverable and citable, the concept of the data paper was introduced in the early 2000’s by the Ecological Society of America. This concept was brought to the attention of the biodiversity community by Chavan and Penev (2011) with the introduction of a new data paper concept, based on a metadata standard, such as the Ecological Metadata Language, and derived from metadata content stored at large data platforms, in this case the Global Biodiversity Information Facility (GBIF). You can read this article for an in-depth discussion of the topic.
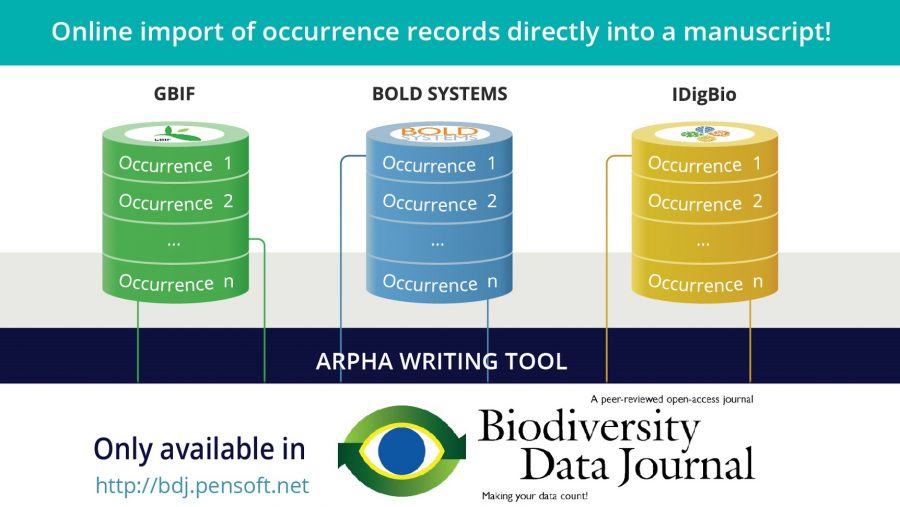
Pensoft’s Biodiversity Data Journal (BDJ) is to the best of our knowledge the first academic journal to have implemented a one-hundred-percent online authoring system for data papers, called ARPHA. Moreover, BDJ and the other Pensoft journals, such as ZooKeys, have already published more than seventy data papers. Therefore, in the remainder of this post we will explain how to use an automated approach to publish a data paper describing an online dataset in Biodiversity Data Journal. The ARPHA system will convert the metadata describing your dataset into a manuscript for you after reading in the metadata! We will illustrate the workflow on the previously mentioned DataONE and GBIF.
The Data Observation Network for Earth (DataONE) is a distributed cyberinfrastructure funded by the U.S. National Science Foundation. It links together over twenty five nodes, primarily in the U.S., hosting biodiversity and biodiversity-related data, and provides an interface to search for data in all of them.
Since butterflies are neat, let’s search for datasets about butterflies at DataONE! Type “Lepidoptera” in the search field and scroll down to the dataset describing “The Effects of Edge Proximity on Butterfly Biodiversity.” You should see something like this:

As you can notice, this resource has two objects associated with it: metadata, which has been highlighted, and the dataset itself. Let’s download the metadata from the cloud! The resulting text file, “Blandy.235.1.xml”, or whatever you want to call it, can be read by humans, but is somewhat cryptic because of all the XML tags. Now, you can import this file into the ARPHA writing platform and the information stored in it would be used to create a data paper! Go to the ARPHA web-site, pwt.pensoft.net, and click on “Start a manuscript,” then scroll all the way down and click on “Import manuscript.”

Upload the “blandy” file and you will see an “Authors’ page,” where you can select which of the authors mentioned in the metadata must be included as authors of the data paper itself. Note that the user of ARPHA uploading the metadata is added to the list of the authors if they is not included in the metadata. After the selection is done, a scholarly article is created by the system with the information from the metadata already in respective sections of the article:

Now, the authors can add some description, edit out errors, tell a story, cite someone – all of this without leaving ARPHA – i.e. do whatever it takes to produce a high-quality scholarly text. After they are done, they can submit their article for peer-review and it could be published in a matter of hours. Voila!
Let’s look at GBIF. Go to “Data -> Explore by country” and select “Saint Vincent and the Grenadines,” an English-speaking Caribbean island. There are, as of the time of writing of this post, 166 occurrence datasets containing data about the islands. Select the dataset from the Museum of Comparative Zoology at Harvard. If you scroll down, you will see the GBIF annotated EML. Download this as a separate text file (if you are in Chrome you can view the source and then use copy-paste). Do the exact same steps as before – go to “Import manuscript” in ARPHA and upload the EML file. The result should be something like this, ready to finalize:

Now, allow us to give a disclaimer here: the authors of this blog post have nothing to do with the two datasets. They have not contributed to any of them, nor do they know the authors. The datasets have been chosen more or less randomly since the authors wanted to demonstrate the functionality with a real-world example. You should only publish data papers if you know the authors or you are the author of the dataset itself. During the actual review process of the paper, the authors that have been included will get an email from the journal!
Having said that, we want to leave you with some caveats and topics for further discussions. Till today, useful and descriptive metadata has not always been present. There are two challenges: metadata completeness and metadata standards. The invention of the EML standard was one of the first efforts to standardize how metadata should be stored in the field of ecology and biodiversity science. Currently, our import system supports the last two versions of the EML standard: 2.1.1 and 2.1.0, but we hope to expand this functionality. In an upcoming version of their search interface, DataONE will provide infographics on the prevalence of the metadata standards at their site (see figure), so there is still work to be done, but if there is a positive feedback from the community, we will definitely keep expanding this feature.

Credit: DataONE
Regarding metadata completeness, our hope is that by enabling scientists to create scholarly papers from their metadata with a single-step process, they will be incentivized to produce high-quality metadata.
This project has received funding from the European Union’s FP7 project EU BON (Building the European Biodiversity Observation Network), grant agreement No 308454, and Horizon 2020 research and innovation project BIG4 (Biosystematics, informatics and genomics of the big 4 insect groups: training tomorrow’s researchers and entrepreneurs) under the Marie Sklodovska-Curie grant agreement No. 542241 for a PhD project titled Technological Implications of the Open Biodiversity Knowledge Management System.