By Mariya Dimitrova, Raïssa Meyer, Pier Luigi Buttigieg, Lyubomir Penev
Data papers are scientific papers which describe a dataset rather than present and discuss research results. The concept was introduced to the biodiversity community by Chavan and Penev in 2011 as the result of a joint project of GBIF and Pensoft.
Since then, Pensoft has implemented the data paper in several of its journals (Fig. 1). The recognition gained through data papers is an important incentive for researchers and data managers to author better quality metadata and to make it Findable, Accessible, Interoperable and Re-usable (FAIR). High quality and FAIRness of (meta)data are promoted through providing peer review, data audit, permanent scientific record and citation credit as for any other scholarly publication. One can read more on the different types of data papers and how they help to achieve these goals in the Strategies and guidelines for scholarly publishing of biodiversity data (https://doi.org/10.3897/rio.3.e12431).
The data paper concept was initially based on the standard metadata descriptions, using the Ecological Metadata Language (EML). Apart from distinguishing a specialised place for dataset descriptions by creating a data paper article type, Pensoft has developed multiple workflows for streamlined import of metadata from various repositories and their conversion into data paper a manuscripts in Pensoft’s ARPHA Writing Tool (AWT). You can read more about the EML workflow in this blogpost.
Similarly, we decided to create a specialised data paper article type for the omics community within Pensoft’s Biodiversity Data Journal to reflect the specific nature of omics data. We established a manuscript template to help standardise the description of such datasets and their most important features. This initiative was supported in part by the IGNITE project.
How can authors publish omics data papers?
There are two ways to do publish omics data papers – (1) to write a data paper manuscript following the respective template in the ARPHA Writing Tool (AWT) or (2) to convert metadata describing a project or study deposited in EMBL-EBI’s European Nucleotide Archive (ENA) into a manuscript within the AWT.
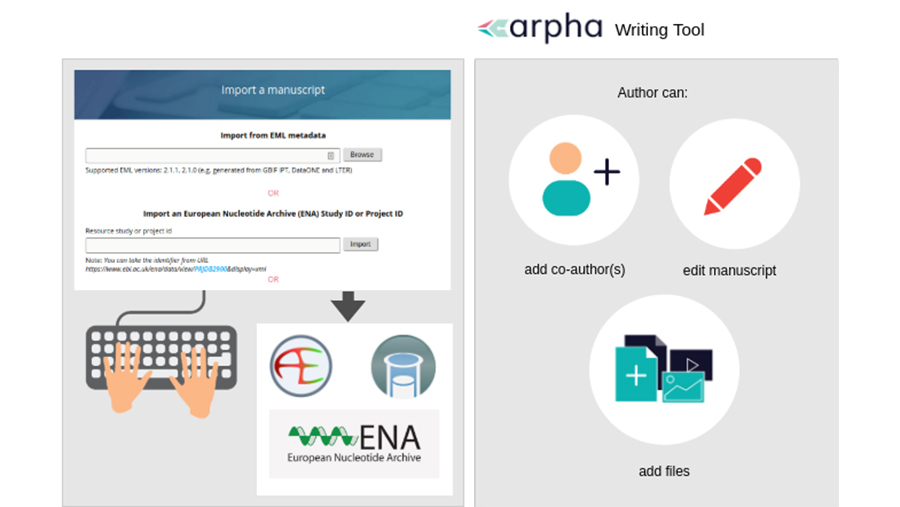
The first method is straightforward but the second one deserves more attention. We focused on metadata published in ENA, which is part of the International Nucleotide Sequence Database Collaboration (INSDC) and synchronises its records with these of the other two members (DDBJ and NCBI). ENA is linked to the ArrayExpress and BioSamples databases, which describe sequencing experiments and samples, and follow the community-accepted metadata standards MINSEQE and MIxS. To auto populate a manuscript with a click of a button, authors can provide the accession number of the relevant ENA Study of Project and our workflow will automatically retrieve all metadata from ENA, as well as any available ArrayExpress or BioSamples records linked to it (Fig. 2). After that, authors can edit any of the article sections in the manuscript by filling in the relevant template fields or creating new sections, adding text, figures, citations and so on.
An important component of the OMICS data paper manuscript is a supplementary table containing MIxS-compliant metadata imported from BioSamples. When available, BioSamples metadata is automatically converted to a long table format and attached to the manuscript. The authors are not permitted to edit or delete it inside the ARPHA Writing Tool. Instead, if desired, they should correct the associated records in the sourced BioSamples database. We have implemented a feature allowing the automatic re-import of corrected BioSamples records inside the supplementary table. In this way, we ensure data integrity and provide a reliable and trusted source for accessing these metadata.
Here is a step-by-step guide for conversion of ENA metadata into a data paper manuscript:
- The author has published a dataset to any of the INSDC databases. They copy its ENA Study or Project accession number.
- The author goes to the Biodiversity Data Journal (BDJ) webpage, clicks the “Start a manuscript” buttоn and selects OMICS Data Paper template in the ARPHA Writing Tool (AWT). Alternatively, the author can also start from the AWT website, click “Create a manuscript”, and select “OMICS Data Paper” as the article type, the Biodiversity Data Journal will be automatically marked by the system. The author clicks the “Import a manuscript” button at the bottom of the webpage.
- The author pastes the ENA Study or Project accession number inside the relevant text box (“Import an European Nucleotide Archive (ENA) Study ID or Project ID”) and clicks “Import”.
- The Project or Study metadata is converted into an OMICS data paper manuscript along with the metadata from ArrayExpress and BioSamples if available. The author can start making changes to the manuscript, invite co-authors and then submit it for technical evaluation, peer review and publication.
For a detailed description of authoring an OMICS data paper, please refer to the Author Guidelines: https://bdj.pensoft.net/about#OmicsDataPapers
Our innovative workflow makes authoring omics data papers much easier and saves authors time and efforts when inserting metadata into the manuscript. It takes advantage of existing links between data repositories to unify biodiversity and omics knowledge into a single narrative. This workflow demonstrates the importance of standardisation and interoperability to integrate data and metadata from different scientific fields.
We have established a special collection for OMICS data papers in the Biodiversity Data Journal. Authors are invited to describe their omics datasets by using the novel streamlined workflow for creating a manuscript at a click of a button from metadata deposited in ENA or by following the template to create their manuscript via the non-automated route.
To stimulate omics data paper publishing, the first 10 papers will be published free of charge. Upon submission of an omics data paper manuscript, do not forget to assign it to the collection Next-generation publishing of omics data.